Liquid crystals (LC) represent a thermodynamically stable state of matter featuring long-range orientational order but complete or partial absence of long-range positional order. This endows LC with anisotropic optical and mechanical properties, thus allowing them to be classified between isotropic liquids and crystalline solids. Dispersion of short double-stranded DNA (dsDNA) in aqueous solutions is a well-known system that exhibits multiple LLC ordered phases at sufficiently high DNA concentrations [5]. The main reasons are rooted in the DNA’s characteristic base stacking and helical architecture, which confer dsDNA molecules with shape anisotropy, chirality and an unusual stiffness with a persistence length Ɩ_p^ssDNA~2nm of about 50 nm (~150 base pairs).
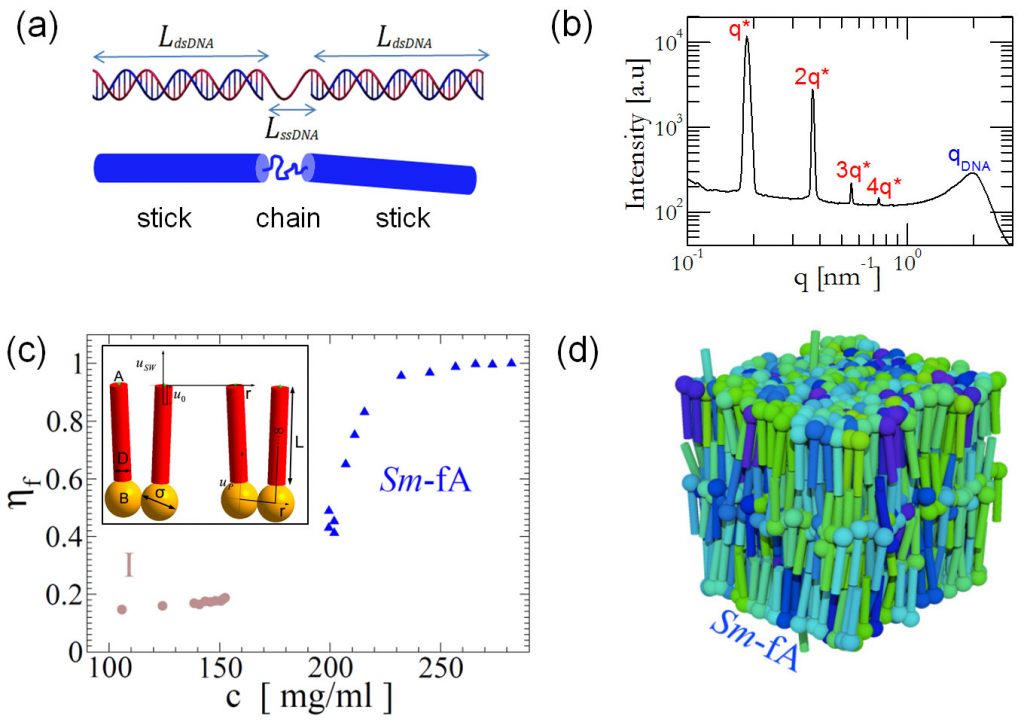
In the present study, we exploit the large difference in the persistence length between dsDNA and single-stranded DNA (ssDNA, Ɩ_p^ssDNA~2nm) in order to construct an all-DNA chain-stick molecule, consisting of a gapped DNA (G-DNA) duplex – with a contour length near the dsDNA’s persistence length – where two stiff blunt-ended dsDNA segments of equal length are connected by an ssDNA flexible spacer (Fig. (a)). Through a combined experimental (Fig. (a, b), title image of this newsletter) and numerical study (Fig. (c, d)), we provide unambiguous evidence of the formation of a smectic-A phase in the aqueous suspensions of all-DNA chain-sticks (Fig. (b, c)).
Surprisingly, our results reveal that the gapped duplexes attain a folded conformation in this two-dimensional fluid layers mesophase, with the vertically aligned molecules having a long molecular axis almost perpendicular to the layer plane (Fig. (c, d)). The good agreement between numerical and experimental results enables us to identify the mechanisms which are key to the formation and thermodynamic stability of this novel LLC phase, which we designate as a “smectic-fA phase” (where the “f” stands for “folded”). We argue that the new liquid crystal phase formed in these all-DNA chain-sticks is a combined result of an entropy-driven (excluded volume reduction) self-assembly and the presence of weak attractive stacking interactions between the duplexes’ blunt-terminal ends.
Our study shows that DNA, as a building block, can offer an exquisitely tunable means to engineer a potentially rich assortment of LLC that can be precisely designed on the sub-nanometer level.
References:
[1] J. D. Watson and F. H. Crick, Nature 171, 737 (1953).
[2] N. C. Seeman, J. Theor. Biol. 99, 237 (1982).
[3] N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater. 3, 17068 (2017).
[4] M. Salamonczyk et al., Nat. Commun. 7, 133358 (2016).
[5] K. Merchant and R. L. Rill, Biophys. J. 73, 3154-3163 (1997).