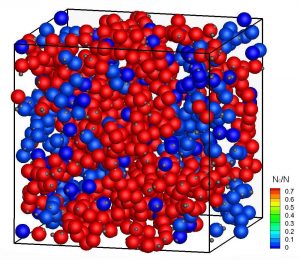
Large, temporary clusters lead to a dramatic slowdown in diffusion for patchy proteins. Copyright: Universität Lund




Large, temporary clusters lead to a dramatic slowdown in diffusion for patchy proteins. Copyright: Universität Lund
Protein diffusion strongly influences numerous processes in biological cells, such as signal transmission. In the dense environment of the cell cytoplasm, protein contacts are omnipresent and individual proteins experience interactions with all the surrounding proteins. While attempts have been made previously to use analogies to colloids in order to model crowding effects, the complexity of protein-protein interactions and the need to measure protein diffusion over length scales comparable to the nearest-neighbour distance represent major obstacles for our understanding of the short-time dynamics in these systems. We have used quasielastic neutron scattering experiments combined with computer simulations to obtain quantitative information about the short-time diffusion of two well-characterized and highly stable bovine eye lens proteins with repulsive and weakly attractive interactions, respectively, in crowded solutions. While diffusion slows down with increasing concentration for both proteins, we find a decrease of the short-time diffusion coefficient for the protein with attractions of almost three orders of magnitude. Supported by computer simulations of colloid-like protein models, we attribute these drastic changes to specific, anisotropic, patchy shortrange protein-protein interactions, which ultimately lead to the formation of large-scale temporal structures. The study provides new insight into emergent patterns of proteins triggered by specific, but typical, weak interactions. It implies that traditional in vitro experiments used to investigate specific protein interactions, recognition processes, and diffusion of proteins under dilute conditions, have to be considered with great caution when trying to understand processes in living cells.
Read more: Bucciarelli S et al., Sci. Adv. 2016;2:e1601432
SoftComp partner: Univ. Lund, ILL, FZJ-Richter, FZJ-Gompper