by Gustavo Bodelón (University of Vigo (UoV), Spain, gbodelon@uvigo.es) & Luis M. Liz-Marzán (UoV, CIC biomaGUNE, Ikerbasque, CIBER-BBN, Spain, llizmarzan@cicbiomagune.es)
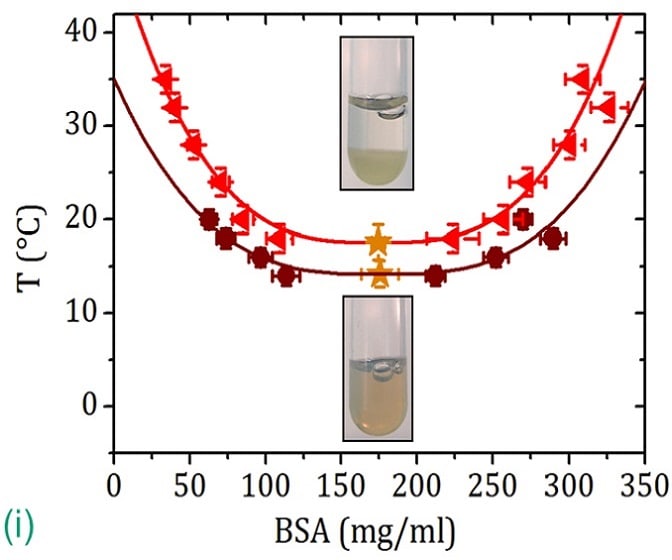
Proteins as Soft Matter: Interactions, Phase Behaviour and Health Research
3rd July 2017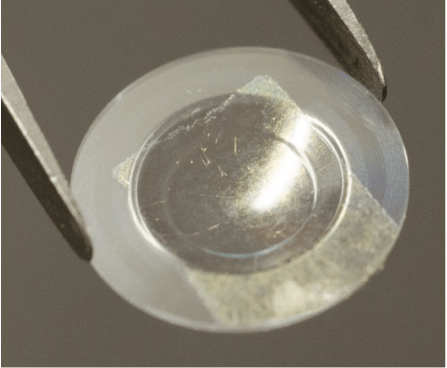
From Novel Soft Materials to Switchable Contact Lenses
5th July 2017
Proteins as Soft Matter: Interactions, Phase Behaviour and Health Research
3rd July 2017
From Novel Soft Materials to Switchable Contact Lenses
5th July 2017The development of non-invasive methods for monitoring chemical communication in bacterial populations is important to provide fundamental insights into these processes as well as the necessary knowledge to manipulate these systems for applications in medicine, drug discovery and biotechnology. Here we describe the application of plasmonic hybrid materials as tools for the detection and imaging of quorum sensing (QS)-derived pyocyanin in live biofilms and in small aggregates of Pseudomonas aeruginosa bacteria, using surface-enhanced resonance Raman scattering (SERRS) spectroscopy. Our work [1] opens new avenues for the application of plasmonic materials as high-performance analytical sensors in microbiology research and in clinical and industrial settings. In addition, bacterial detection through plasmonics-based ultrasensitive detection could aid in the development of more effective strategies for prevention and control of such microorganisms.
QS is a cell-to-cell communication process mediated by small diffusible signalling molecules, which allows bacterial populations to synchronize gene expression in a cell density-dependent manner. QS-regulated genes are associated with collective responses such as antibiotic production, virulence or biofilm formation, all of which are important for bacterial survival and pathogenicity [2]. Thus, understanding QS communication could provide us with a powerful means of antagonizing harmful microbial strains and potentiating beneficial ones [3]. Biofilms are microbial sessile populations characterized by densely packed aggregates of cells embedded in self-produced extracellular polymer substances [4]. Bacterial biofilms are behind industrial biofouling and biocorrosion, and represent a threat to human health, as they are intimately linked with the pathogenesis of acute and persistent infectious diseases [5]. In addition, the high cell density within biofilms provides an optimal environment for intercellular communication processes, including QS.
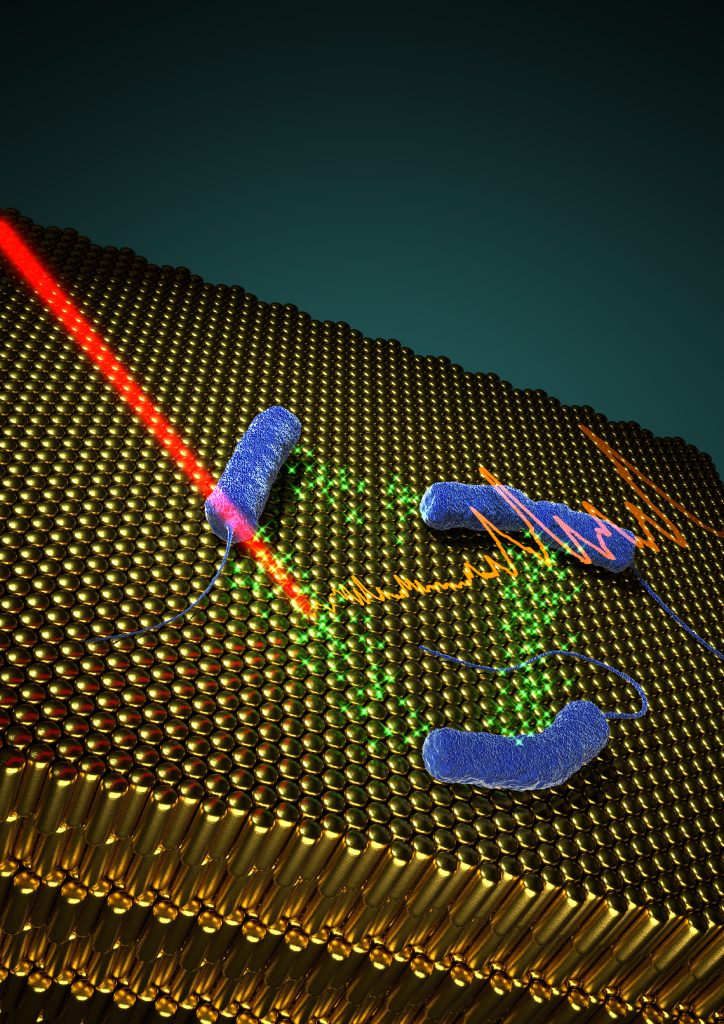
In situ SERRS detection of quorum sensing pyocyanin in cultures of P. aeruginosa bacteria grown on plasmonic nanostructured materials.
Copyright: Andrea La Porta

Gustavo Bodelón, University of Vigo (UoV), Spain
Copyright: private

Luis M. Liz-Marzán, UoV, CIC biomaGUNE, Ikerbasque, CIBER-BBN, Spain
Copyright: private
We focused this work on the detection of pyocyanin, a QS-signalling molecule of P. aeruginosa that plays important roles in biofilm morphogenesis in this important human pathogen [1]. To this end, we fabricated hybrid materials comprising a plasmonic component embedded in a porous matrix, as multifunctional platforms to develop biofilms of P. aeruginosa, and simultaneously to detect pyocyanin by SERRS non-invasively (Fig.). The porous nature of the substrates plays an important role in restricting the contact of the plasmonic component with biomolecules that could contaminate the SERRS signal and thereby hinder the selectivity and sensitivity of the assay. The most efficient substrates comprise gold nanorod supercrystals (schematically shown in the figure), covered by mesoporous silica, which allowed us to detect pyocyanin at early growth times, during the formation of biofilms. Alternatively, the use of nanoparticle-doped hydrogels enabled detection in subcutaneous implants. Owing to the high enhancement factors of the plasmonic substrates, ultrasensitive molecular detection could be achieved, down to 10-15 M.
This approach, combining purpose-designed nanostructured hybrid materials and SERRS, demonstrates the potential of plasmonics for in situ, label-free monitoring of biological processes mediated by SERRS-active biomolecules (e.g. QS communication) in live microbial populations. It paves the way toward the assessment of harmful microbial contamination in medicine (e.g. catheters, implantable medical devices) and in the food industry.
References:
[1] Bodelon G et al. Nat. Mater. 2016;15(11):1203-1211.
[2] Papenfort K et al. Nat. Rev. Microbiol. 2016;14(9):576-588.
[3] LaSarre B et al. Microbiol. Mol. Biol. Rev. 2013;77:73-111.
[4] Hall-Stoodley L et al. Nat. Rev. Microbiol. 2004;2:95-108.
[5] Rybtke M at al. J. Mol. Biol. 2015;427(23):3628-45.
Note: This article was first published in July 2017 in the SoftComp Newsletter Issue 14.
