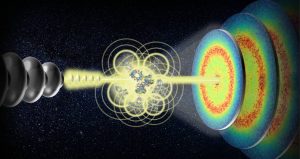
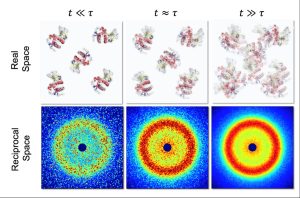
by Frank Schreiber, frank.schreiber@uni-tuebingen.de, Institute of Applied Physics of University of Tübingen, Germany, on behalf of the entire collaboration [*]
Novel experiment opens up possibility of studying collective dynamics providing insight for better drug design.
Abstract
Proteins carry out a wide range of important roles in our bodies. Knowledge about how they move around and through the crowded environments of our cells and tissues, is key to understanding how they carry out these crucial functions. Experiments exploring protein dynamics however, often use samples that do not reflect the crowded and complex environments of the cell. At the MID instrument at European XFEL, scientists carried out X-ray photon correlation spectroscopy experiments to measure nanoscopic structural dynamics of protein solution samples. The combination of this technique with the high repetition rate of X-ray pulses generated by European XFEL opens up the unique possibility of studying collective dynamics in protein solutions with high concentration; particularly interesting in the context of intracellular transport in eukaryotic cells, and drug design.
Catalysts, transporters, gate-keepers, messengers – proteins carry out a vast range of different roles in our bodies. In addition to their structure, how they move through, and interact and react with their crowded and complex environment impacts their function. And yet, our understanding of the dynamics of processes such as protein diffusion and transport in cells and tissues is still lacking.
Protein dynamics in crowded media, especially diffusion and transport, are essential for biological processes and cellular function [1]. A comprehensive knowledge of how proteins behave in these environments is especially useful for example for the design of new drugs. When drugs enter our blood streams, it is the proteins that bind them and transport and distribute them around the body XFEL. How fast this happens, for example, is crucial information for understanding how long it takes for a drug to be dispersed, but also, in the case of for example cancer drugs, how and when toxicity levels begin to become too much as the amount of protein bound drug increases in the bloodstream. A comprehensive understanding of dynamics of these kinds of processes could inform more effective treatments with fewer side effects for example.