Proteins occur in nature in enormous numbers and fulfil a huge range of biological tasks. Yet they are all simply made up of the same 20 amino acid building blocks, assembled into polymers and folded into a wide variety of shapes that determine their function. Proteins should however not be considered as a single atomic arrangement: they are flexible, exploring a continuous conformational space often described as a set of low energy states connected by higher energy transition paths. Current experimental techniques provide a good picture of the most stable conformations, but little on the transition path or intermediate states. Computational techniques such as molecular dynamics simulations can be used to characterize protein dynamics by sampling their conformational space. However, the odds a simulation will sample a specific conformation are inversely proportional to its energy, making the discovery of transition states a rare event.
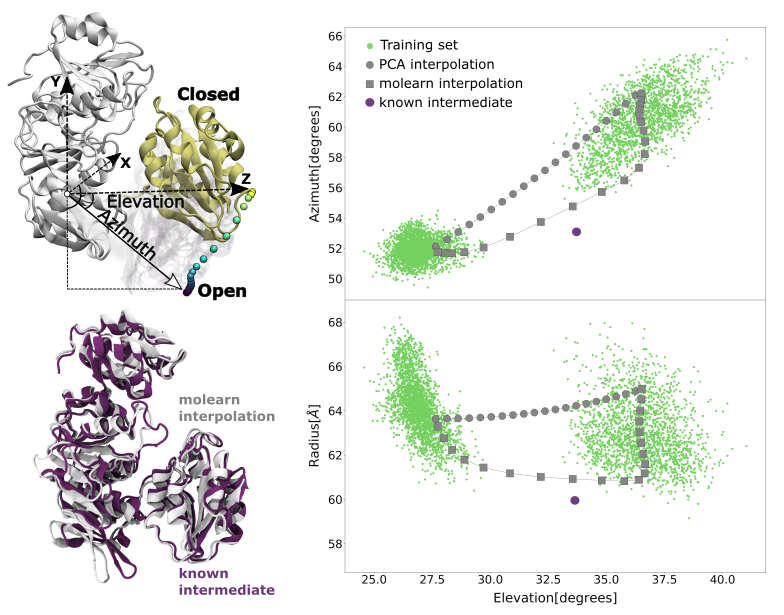
Given the remarkable success of deep generative neural networks in generating believable synthetic images, videos and texts, we designed a neural network that, trained with a discrete set of structures produced via molecular simulations, can generate protein conformations. To ensure the network produces structures respecting physical laws, we also designed a new training procedure whereby we penalise high-energy conformations generated outside of the sampled space. As a result, our network attempts to generate transition paths of minimal energy. To demonstrate the usefulness of our approach, called molearn, we successfully challenged it with the prediction of the transition path between known conformations of the bacterial protein MurD.
Free access to molearn : www.github.com/degiacom/molearn
Read more:
Ramaswamy V.K. et al.,
Phys. Rev. X 11, 011052 (2021)
SoftComp partner:
Durham University